Calculating band indices 
Sign up to the DEA Sandbox to run this notebook interactively from a browser
Compatibility: Notebook currently compatible with both the
DEA SandboxandNCIenvironmentsProducts used: ga_ls8c_ard_3
Background
Remote sensing indices are combinations of spectral bands used to highlight features in the data and the underlying landscape. Using Digital Earth Australia’s archive of analysis-ready satellite data, we can easily calculate a wide range of remote sensing indices that can be used to assist in mapping and monitoring features like vegetation and water consistently through time, or as inputs to machine learning or classification algorithms.
Description
This notebook demonstrates how to:
Calculate an index manually using
xarrayCalculate one or multiple indices using the function
calculate_indicesfromdea_tools.bandindicesCalculate indices while dropping original bands from a dataset
Calculate an index in-place without duplicating original data to save memory on large datasets
Getting started
To run this analysis, run all the cells in the notebook, starting with the “Load packages” cell.
Load packages
[1]:
%matplotlib inline
import datacube
import matplotlib.pyplot as plt
import sys
sys.path.insert(1, '../Tools/')
from dea_tools.datahandling import load_ard
from dea_tools.bandindices import calculate_indices
from dea_tools.plotting import rgb
Connect to the datacube
[2]:
dc = datacube.Datacube(app='Calculating_band_indices')
Create a query and load satellite data
To demonstrate how to compute a remote sensing index, we first need to load in a time series of satellite data for an area. We will use data from the Landsat 8 satellite:
[3]:
# Create a reusable query
query = {
'x': (149.06, 149.17),
'y': (-35.27, -35.32),
'time': ('2020-01', '2020-12'),
'measurements': [
'nbart_blue', 'nbart_green', 'nbart_red', 'nbart_nir', 'nbart_swir_1',
'nbart_swir_2'
],
'output_crs': 'EPSG:3577',
'resolution': (-30, 30),
'group_by': 'solar_day'
}
# Load available data from Landsat 8 and filter to retain only times
# with at least 99% good data
ds = load_ard(dc=dc, products=['ga_ls8c_ard_3'], min_gooddata=0.99, **query)
Finding datasets
ga_ls8c_ard_3
Counting good quality pixels for each time step using fmask
/env/lib/python3.10/site-packages/rasterio/warp.py:387: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
dest = _reproject(
Filtering to 5 out of 44 time steps with at least 99.0% good quality pixels
Applying fmask pixel quality/cloud mask
Loading 5 time steps
Plot the first image to see what our area looks like
We can use the rgb function to plot the first timestep in our dataset as a true colour RGB image:
[4]:
# Plot as an RGB image
rgb(ds, index=0, size=4);

Calculate an index for this area manually
One of the most commonly used remote sensing indices is the Normalised Difference Vegetation Index or NDVI. This index uses the ratio of the red and near-infrared (NIR) bands to identify live green vegetation. The formula for NDVI is:
When interpreting this index, high values indicate vegetation, and low values indicate soil or water.
[5]:
# Calculate NDVI using the formula above
ds['ndvi'] = (ds.nbart_nir - ds.nbart_red) / (ds.nbart_nir + ds.nbart_red)
# Plot the results for one time step to see what they look like:
ds.ndvi.isel(time=0).plot(vmin=0, vmax=1, cmap='gist_earth_r');
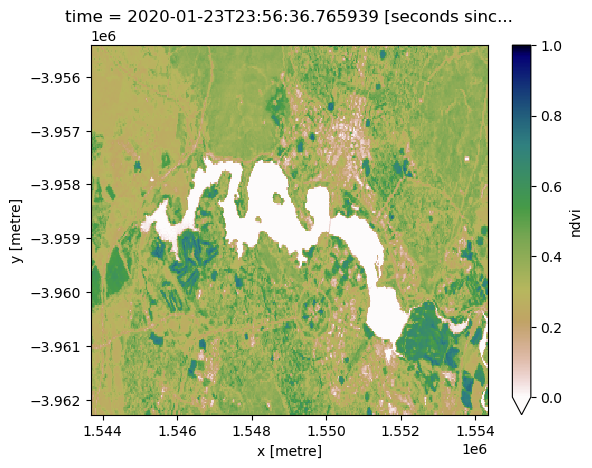
In the image above, vegetation shows up as green (NDVI > 0). Sand shows up as yellow (NDVI ~ 0) and water shows up as red (NDVI < 0).
Calculate an index for the same area using calculate_indices function
The calculate_indices function provides an easier way to calculate a wide range of remote sensing indices, including:
AWEI_ns (Automated Water Extraction Index,no shadows, Feyisa 2014)
AWEI_sh (Automated Water Extraction Index,shadows, Feyisa 2014)
BAEI (Built-Up Area Extraction Index, Bouzekri et al. 2015)
BAI (Burn Area Index, Martin 1998)
BSI (Bare Soil Index, Rikimaru et al. 2002)
BUI (Built-Up Index, He et al. 2010)
CMR (Clay Minerals Ratio, Drury 1987)
EVI (Enhanced Vegetation Index, Huete 2002)
FMR (Ferrous Minerals Ratio, Segal 1982)
IOR (Iron Oxide Ratio, Segal 1982)
LAI (Leaf Area Index, Boegh 2002)
MNDWI (Modified Normalised Difference Water Index, Xu 1996)
MSAVI (Modified Soil Adjusted Vegetation Index, Qi et al. 1994)
NBI (New Built-Up Index, Jieli et al. 2010)
NBR (Normalised Burn Ratio, Lopez Garcia 1991)
NDBI (Normalised Difference Built-Up Index, Zha 2003)
NDCI (Normalised Difference Chlorophyll Index, Mishra & Mishra, 2012)
NDMI (Normalised Difference Moisture Index, Gao 1996)
NDSI (Normalised Difference Snow Index, Hall 1995)
NDVI (Normalised Difference Vegetation Index, Rouse 1973)
NDWI (Normalised Difference Water Index, McFeeters 1996)
NIRv (Near-Infrared Reflectance of Vegetation, Badgley et al. 2017)
kNDVI (Kernel Normalized Difference Vegetation Index, Camps-Valls et al. 2021)
SAVI (Soil Adjusted Vegetation Index, Huete 1988)
TCB (Tasseled Cap Brightness, Crist 1985)
TCG (Tasseled Cap Greeness, Crist 1985)
TCW (Tasseled Cap Wetness, Crist 1985)
WI (Water Index, Fisher 2016)
Using calculate_indices, we get the same result:
[6]:
# Calculate NDVI using `calculate indices`
ds_ndvi = calculate_indices(ds, index='NDVI', collection='ga_ls_3')
# Plot the results
ds_ndvi.NDVI.isel(time=0).plot(vmin=0, vmax=1, cmap='gist_earth_r');
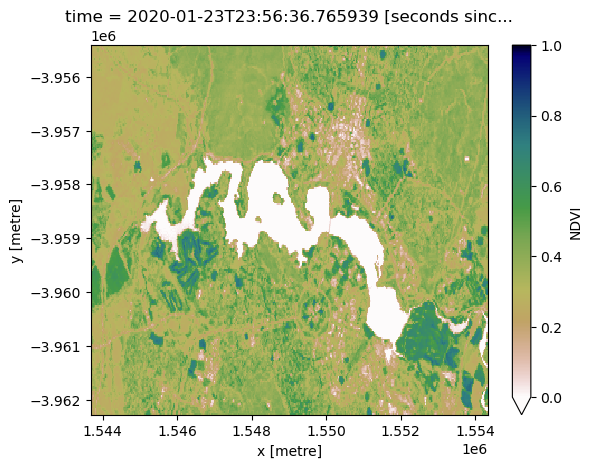
Note: when using the
calculate_indicesfunction, it is important to set thecollectionparameter correctly. This is because different satellite collections use different names for the same bands, which can lead to invalid results if not accounted for.
For Landsat (i.e. GA Landsat Collection 3), specify
collection='ga_ls_3'.For Sentinel 2 (i.e. GA Sentinel 2 Collection 3), specify
collection='ga_s2_3'.For Landsat geomedian products, specify
collection='ga_gm_3'.
Using calculate_indices to calculate multiple indices at once
The calculate_indices function makes it straightforward to calculate multiple remote sensing indices in one line of code.
In the example below, we will calculate NDVI as well as two common water indices: the Normalised Difference Water Index (NDWI), and the Modified Normalised Difference Index (MNDWI). The new indices will appear in the list of data_variables below:
[7]:
# Calculate multiple indices
ds_multi = calculate_indices(ds, index=['NDVI', 'NDWI', 'MNDWI'], collection='ga_ls_3')
ds_multi
[7]:
<xarray.Dataset> Size: 16MB
Dimensions: (time: 5, y: 229, x: 355)
Coordinates:
* time (time) datetime64[ns] 40B 2020-01-23T23:56:36.765939 ... 20...
* y (y) float64 2kB -3.955e+06 -3.955e+06 ... -3.962e+06
* x (x) float64 3kB 1.544e+06 1.544e+06 ... 1.554e+06 1.554e+06
spatial_ref int32 4B 3577
Data variables:
nbart_blue (time, y, x) float32 2MB 716.0 731.0 741.0 ... 368.0 308.0
nbart_green (time, y, x) float32 2MB 1.015e+03 1.031e+03 ... 689.0 583.0
nbart_red (time, y, x) float32 2MB 1.371e+03 1.395e+03 ... 666.0 545.0
nbart_nir (time, y, x) float32 2MB 2.327e+03 2.334e+03 ... 2.788e+03
nbart_swir_1 (time, y, x) float32 2MB 3.687e+03 3.787e+03 ... 1.347e+03
nbart_swir_2 (time, y, x) float32 2MB 2.639e+03 2.738e+03 ... 751.0
ndvi (time, y, x) float32 2MB 0.2585 0.2518 0.2632 ... 0.6612 0.673
NDVI (time, y, x) float32 2MB 0.2585 0.2518 0.2632 ... 0.6612 0.673
NDWI (time, y, x) float32 2MB -0.3926 -0.3872 ... -0.6515 -0.6541
MNDWI (time, y, x) float32 2MB -0.5683 -0.572 ... -0.449 -0.3959
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refDropping original bands from a dataset
We can also drop the original satellite bands from the dataset using drop=True. The dataset produced below should now only include the new 'NDVI', 'NDWI', 'MNDWI' bands under data_variables:
[8]:
# Calculate multiple indices and drop original bands
ds_drop = calculate_indices(ds,
index=['NDVI', 'NDWI', 'MNDWI'],
drop=True,
collection='ga_ls_3')
ds_drop
Dropping bands ['nbart_blue', 'nbart_green', 'nbart_red', 'nbart_nir', 'nbart_swir_1', 'nbart_swir_2', 'ndvi']
[8]:
<xarray.Dataset> Size: 5MB
Dimensions: (time: 5, y: 229, x: 355)
Coordinates:
* time (time) datetime64[ns] 40B 2020-01-23T23:56:36.765939 ... 202...
* y (y) float64 2kB -3.955e+06 -3.955e+06 ... -3.962e+06 -3.962e+06
* x (x) float64 3kB 1.544e+06 1.544e+06 ... 1.554e+06 1.554e+06
spatial_ref int32 4B 3577
Data variables:
NDVI (time, y, x) float32 2MB 0.2585 0.2518 0.2632 ... 0.6612 0.673
NDWI (time, y, x) float32 2MB -0.3926 -0.3872 ... -0.6515 -0.6541
MNDWI (time, y, x) float32 2MB -0.5683 -0.572 ... -0.449 -0.3959
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refCalculating indices in-place to reduce memory usage for large datasets
By default, the calculate_indices function will create a new copy of the original data that contains the newly generated remote sensing indices. This can be problematic for large datasets, as this effectively doubles the amount of data that is stored in memory.
To calculate remote sensing indices directly in-place within the original dataset without copying the data, we can run the function with the parameter inplace=True. Note that we don’t need to assign any output for the function, as the changes will be made to the original data.
[9]:
# Calculate index in place without copying data
calculate_indices(ds,
index=['TCW'],
collection='ga_ls_3',
inplace=True)
ds
[9]:
<xarray.Dataset> Size: 13MB
Dimensions: (time: 5, y: 229, x: 355)
Coordinates:
* time (time) datetime64[ns] 40B 2020-01-23T23:56:36.765939 ... 20...
* y (y) float64 2kB -3.955e+06 -3.955e+06 ... -3.962e+06
* x (x) float64 3kB 1.544e+06 1.544e+06 ... 1.554e+06 1.554e+06
spatial_ref int32 4B 3577
Data variables:
nbart_blue (time, y, x) float32 2MB 716.0 731.0 741.0 ... 368.0 308.0
nbart_green (time, y, x) float32 2MB 1.015e+03 1.031e+03 ... 689.0 583.0
nbart_red (time, y, x) float32 2MB 1.371e+03 1.395e+03 ... 666.0 545.0
nbart_nir (time, y, x) float32 2MB 2.327e+03 2.334e+03 ... 2.788e+03
nbart_swir_1 (time, y, x) float32 2MB 3.687e+03 3.787e+03 ... 1.347e+03
nbart_swir_2 (time, y, x) float32 2MB 2.639e+03 2.738e+03 ... 751.0
ndvi (time, y, x) float32 2MB 0.2585 0.2518 0.2632 ... 0.6612 0.673
TCW (time, y, x) float32 2MB -0.3098 -0.3214 ... -0.09681 -0.06346
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refAdditional information
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Australia data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Discord chat or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
GitHub.
Last modified: September 2025
Compatible datacube version:
[10]:
print(datacube.__version__)
1.8.19
Tags
Tags: sandbox compatible, NCI compatible, load_ard, rgb, calculate_indices, landsat 8, index calculation, NDVI, NDWI, MNDWI