Working with time in Xarray 
Sign up to the DEA Sandbox to run this notebook interactively from a browser
Compatibility: Notebook currently compatible with both the
NCIandDEA SandboxenvironmentsProducts used: ga_ls8c_ard_3
Description
The xarray Python package provides many useful techniques for dealing with time series data that can be applied to Digital Earth Australia data. This notebook demonstrates how to use xarray techniques to:
Select different time periods of data (e.g. year, month, day) from an
xarray.DatasetUsing datetime accessors to extract additional information from a dataset’s
timedimensionSummarising time series data for different time periods using
.groupby()and.resample()Interpolating time series data to estimate landscape conditions at a specific date that the satellite did not observe
For additional information about the techniques demonstrated below, refer to the xarray Time series data guide.
Getting started
To run this analysis, run all the cells in the notebook, starting with the “Load packages” cell.
Load packages
[1]:
%matplotlib inline
import datacube
import matplotlib.pyplot as plt
Connect to the datacube
[2]:
dc = datacube.Datacube(app='Working_with_time')
Loading Landsat data
First, we load in a two years of Landsat 8 data, and apply a simple cloud mask. To learn more about masking data, see the masking data notebook.
[3]:
# Set up a location for the analysis
query = {
'x': (142.41, 142.51),
'y': (-32.22, -32.32),
'time': ('2015-01-01', '2016-12-31'),
'measurements': ['nbart_nir', 'fmask'],
'output_crs': 'EPSG:3577',
'resolution': (-30, 30)
}
# Load Landsat 8 data
ds = dc.load(product='ga_ls8c_ard_3', group_by='solar_day', **query)
# Apply simple cloud mask (keep only clear, water or snow observations)
ds = ds.where(ds.fmask.isin([1, 4, 5]))
print(ds)
<xarray.Dataset>
Dimensions: (time: 46, x: 342, y: 398)
Coordinates:
* time (time) datetime64[ns] 2015-01-06T00:20:26.943424 ... 2016-12...
* y (y) float64 -3.552e+06 -3.552e+06 ... -3.563e+06 -3.563e+06
* x (x) float64 9.709e+05 9.71e+05 9.71e+05 ... 9.811e+05 9.812e+05
spatial_ref int32 3577
Data variables:
nbart_nir (time, y, x) float64 nan nan nan nan nan ... nan nan nan nan
fmask (time, y, x) float64 nan nan nan nan nan ... nan nan nan nan
Attributes:
crs: EPSG:3577
grid_mapping: spatial_ref
Working with time
Indexing by time
We can select data for an entire year by passing a string to .sel():
[4]:
ds.sel(time='2015')
[4]:
<xarray.Dataset>
Dimensions: (time: 23, x: 342, y: 398)
Coordinates:
* time (time) datetime64[ns] 2015-01-06T00:20:26.943424 ... 2015-12...
* y (y) float64 -3.552e+06 -3.552e+06 ... -3.563e+06 -3.563e+06
* x (x) float64 9.709e+05 9.71e+05 9.71e+05 ... 9.811e+05 9.812e+05
spatial_ref int32 3577
Data variables:
nbart_nir (time, y, x) float64 nan nan nan ... 3.123e+03 3.148e+03
fmask (time, y, x) float64 nan nan nan nan nan ... 1.0 1.0 1.0 1.0
Attributes:
crs: EPSG:3577
grid_mapping: spatial_ref- time: 23
- x: 342
- y: 398
- time(time)datetime64[ns]2015-01-06T00:20:26.943424 ... 2...
- units :
- seconds since 1970-01-01 00:00:00
array(['2015-01-06T00:20:26.943424000', '2015-01-22T00:20:21.686938000', '2015-02-07T00:20:18.471354000', '2015-02-23T00:20:12.433781000', '2015-03-11T00:20:02.105880000', '2015-03-27T00:19:53.920971000', '2015-04-12T00:19:46.703592000', '2015-04-28T00:19:40.864449000', '2015-05-14T00:19:24.695756000', '2015-05-30T00:19:28.951301000', '2015-06-15T00:19:41.139454000', '2015-07-01T00:19:47.228818000', '2015-07-17T00:19:57.584270000', '2015-08-02T00:20:01.251247000', '2015-08-18T00:20:08.061952000', '2015-09-03T00:20:12.861679000', '2015-09-19T00:20:21.648088000', '2015-10-05T00:20:25.198218000', '2015-10-21T00:20:27.331461000', '2015-11-06T00:20:30.948967000', '2015-11-22T00:20:32.825090000', '2015-12-08T00:20:31.424104000', '2015-12-24T00:20:32.375097000'], dtype='datetime64[ns]') - y(y)float64-3.552e+06 ... -3.563e+06
- units :
- metre
- resolution :
- -30.0
- crs :
- EPSG:3577
array([-3551565., -3551595., -3551625., ..., -3563415., -3563445., -3563475.])
- x(x)float649.709e+05 9.71e+05 ... 9.812e+05
- units :
- metre
- resolution :
- 30.0
- crs :
- EPSG:3577
array([970935., 970965., 970995., ..., 981105., 981135., 981165.])
- spatial_ref()int323577
- spatial_ref :
- PROJCS["GDA94 / Australian Albers",GEOGCS["GDA94",DATUM["Geocentric_Datum_of_Australia_1994",SPHEROID["GRS 1980",6378137,298.257222101,AUTHORITY["EPSG","7019"]],AUTHORITY["EPSG","6283"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4283"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["latitude_of_center",0],PARAMETER["longitude_of_center",132],PARAMETER["standard_parallel_1",-18],PARAMETER["standard_parallel_2",-36],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","3577"]]
- grid_mapping_name :
- albers_conical_equal_area
array(3577, dtype=int32)
- nbart_nir(time, y, x)float64nan nan nan ... 3.123e+03 3.148e+03
- units :
- 1
- nodata :
- -999
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[ nan, nan, nan, ..., 3442., 3250., 2588.], [ nan, nan, nan, ..., 3635., 3277., 2530.], [ nan, nan, nan, ..., 3582., 3469., 3057.], ..., [1856., 1853., 1940., ..., 3208., 3886., 3881.], [1909., 1881., 1903., ..., 3059., 3376., 3467.], [1888., 1928., 1994., ..., 2561., 3200., 3317.]], [[1928., 2245., 2262., ..., 2810., 2831., 2407.], [1830., 2065., 2443., ..., 2837., 2938., 2454.], [1835., 1883., 2161., ..., 2971., 2901., 2536.], ..., [1742., 1756., 1778., ..., 3228., 3865., 3816.], [1803., 1813., 1828., ..., 2932., 3370., 3429.], [1698., 1760., 1870., ..., 2658., 3314., 3358.]], [[2023., 2310., 2287., ..., 2827., 2903., 2464.], [1901., 2193., 2424., ..., 2932., 2870., 2580.], [1947., 1964., 2352., ..., 3061., 2911., 2658.], ..., ... ..., [2069., 2083., 2236., ..., 3394., 3887., 3863.], [2069., 2143., 2252., ..., 3073., 3493., 3451.], [2196., 2337., 2358., ..., 2740., 3324., 3228.]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [2363., 2485., 2545., ..., nan, nan, nan], [2374., 2444., 2579., ..., nan, nan, nan], [2493., 2622., 2671., ..., nan, nan, nan]], [[2135., 2371., 2421., ..., 2799., 2767., 2516.], [2125., 2332., 2566., ..., 2974., 2800., 2576.], [2143., 2226., 2422., ..., 2995., 2967., 2748.], ..., [1992., 2045., 2170., ..., 3259., 3767., 3800.], [2027., 2077., 2204., ..., 3008., 3374., 3356.], [2132., 2298., 2276., ..., 2572., 3123., 3148.]]]) - fmask(time, y, x)float64nan nan nan nan ... 1.0 1.0 1.0 1.0
- units :
- 1
- nodata :
- 0
- flags_definition :
- {'fmask': {'bits': [0, 1, 2, 3, 4, 5, 6, 7], 'values': {'0': 'nodata', '1': 'valid', '2': 'cloud', '3': 'shadow', '4': 'snow', '5': 'water'}, 'description': 'Fmask'}}
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[nan, nan, nan, ..., 1., 1., 1.], [nan, nan, nan, ..., 1., 1., 1.], [nan, nan, nan, ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., ... ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan], [ 1., 1., 1., ..., nan, nan, nan]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]]])
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
Or select a single month:
[5]:
ds.sel(time='2015-05')
[5]:
<xarray.Dataset>
Dimensions: (time: 2, x: 342, y: 398)
Coordinates:
* time (time) datetime64[ns] 2015-05-14T00:19:24.695756 2015-05-30T...
* y (y) float64 -3.552e+06 -3.552e+06 ... -3.563e+06 -3.563e+06
* x (x) float64 9.709e+05 9.71e+05 9.71e+05 ... 9.811e+05 9.812e+05
spatial_ref int32 3577
Data variables:
nbart_nir (time, y, x) float64 2.02e+03 2.276e+03 2.164e+03 ... nan nan
fmask (time, y, x) float64 1.0 1.0 1.0 1.0 1.0 ... nan nan nan nan
Attributes:
crs: EPSG:3577
grid_mapping: spatial_ref- time: 2
- x: 342
- y: 398
- time(time)datetime64[ns]2015-05-14T00:19:24.695756 2015-...
- units :
- seconds since 1970-01-01 00:00:00
array(['2015-05-14T00:19:24.695756000', '2015-05-30T00:19:28.951301000'], dtype='datetime64[ns]') - y(y)float64-3.552e+06 ... -3.563e+06
- units :
- metre
- resolution :
- -30.0
- crs :
- EPSG:3577
array([-3551565., -3551595., -3551625., ..., -3563415., -3563445., -3563475.])
- x(x)float649.709e+05 9.71e+05 ... 9.812e+05
- units :
- metre
- resolution :
- 30.0
- crs :
- EPSG:3577
array([970935., 970965., 970995., ..., 981105., 981135., 981165.])
- spatial_ref()int323577
- spatial_ref :
- PROJCS["GDA94 / Australian Albers",GEOGCS["GDA94",DATUM["Geocentric_Datum_of_Australia_1994",SPHEROID["GRS 1980",6378137,298.257222101,AUTHORITY["EPSG","7019"]],AUTHORITY["EPSG","6283"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4283"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["latitude_of_center",0],PARAMETER["longitude_of_center",132],PARAMETER["standard_parallel_1",-18],PARAMETER["standard_parallel_2",-36],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","3577"]]
- grid_mapping_name :
- albers_conical_equal_area
array(3577, dtype=int32)
- nbart_nir(time, y, x)float642.02e+03 2.276e+03 ... nan nan
- units :
- 1
- nodata :
- -999
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[2020., 2276., 2164., ..., 3027., 2811., 2695.], [1820., 2120., 2358., ..., 3058., 2674., 2702.], [1863., 1901., 2214., ..., 3111., 2963., 2868.], ..., [1763., 1775., 1812., ..., 3156., 3853., 3757.], [1745., 1832., 1903., ..., 2850., 3152., 3127.], [2005., 2096., 2138., ..., 2466., 3085., 3122.]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - fmask(time, y, x)float641.0 1.0 1.0 1.0 ... nan nan nan nan
- units :
- 1
- nodata :
- 0
- flags_definition :
- {'fmask': {'bits': [0, 1, 2, 3, 4, 5, 6, 7], 'values': {'0': 'nodata', '1': 'valid', '2': 'cloud', '3': 'shadow', '4': 'snow', '5': 'water'}, 'description': 'Fmask'}}
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]])
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
Or select a range of dates using slice(). This selects all observations between the two dates, inclusive of both the start and stop values:
[6]:
ds.sel(time=slice('2015-06', '2016-01'))
[6]:
<xarray.Dataset>
Dimensions: (time: 15, x: 342, y: 398)
Coordinates:
* time (time) datetime64[ns] 2015-06-15T00:19:41.139454 ... 2016-01...
* y (y) float64 -3.552e+06 -3.552e+06 ... -3.563e+06 -3.563e+06
* x (x) float64 9.709e+05 9.71e+05 9.71e+05 ... 9.811e+05 9.812e+05
spatial_ref int32 3577
Data variables:
nbart_nir (time, y, x) float64 nan nan nan ... 3.324e+03 3.307e+03
fmask (time, y, x) float64 nan nan nan nan nan ... 1.0 1.0 1.0 1.0
Attributes:
crs: EPSG:3577
grid_mapping: spatial_ref- time: 15
- x: 342
- y: 398
- time(time)datetime64[ns]2015-06-15T00:19:41.139454 ... 2...
- units :
- seconds since 1970-01-01 00:00:00
array(['2015-06-15T00:19:41.139454000', '2015-07-01T00:19:47.228818000', '2015-07-17T00:19:57.584270000', '2015-08-02T00:20:01.251247000', '2015-08-18T00:20:08.061952000', '2015-09-03T00:20:12.861679000', '2015-09-19T00:20:21.648088000', '2015-10-05T00:20:25.198218000', '2015-10-21T00:20:27.331461000', '2015-11-06T00:20:30.948967000', '2015-11-22T00:20:32.825090000', '2015-12-08T00:20:31.424104000', '2015-12-24T00:20:32.375097000', '2016-01-09T00:20:27.829692000', '2016-01-25T00:20:28.787852000'], dtype='datetime64[ns]') - y(y)float64-3.552e+06 ... -3.563e+06
- units :
- metre
- resolution :
- -30.0
- crs :
- EPSG:3577
array([-3551565., -3551595., -3551625., ..., -3563415., -3563445., -3563475.])
- x(x)float649.709e+05 9.71e+05 ... 9.812e+05
- units :
- metre
- resolution :
- 30.0
- crs :
- EPSG:3577
array([970935., 970965., 970995., ..., 981105., 981135., 981165.])
- spatial_ref()int323577
- spatial_ref :
- PROJCS["GDA94 / Australian Albers",GEOGCS["GDA94",DATUM["Geocentric_Datum_of_Australia_1994",SPHEROID["GRS 1980",6378137,298.257222101,AUTHORITY["EPSG","7019"]],AUTHORITY["EPSG","6283"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4283"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["latitude_of_center",0],PARAMETER["longitude_of_center",132],PARAMETER["standard_parallel_1",-18],PARAMETER["standard_parallel_2",-36],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","3577"]]
- grid_mapping_name :
- albers_conical_equal_area
array(3577, dtype=int32)
- nbart_nir(time, y, x)float64nan nan nan ... 3.324e+03 3.307e+03
- units :
- 1
- nodata :
- -999
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[1940., 2107., 1954., ..., 2528., 2551., 2408.], [1759., 2062., 2190., ..., 2519., 2611., 2131.], [1801., 1896., 2256., ..., 2704., 2440., 2221.], ..., [1513., 1528., 1522., ..., 2651., 3199., 3184.], [1561., 1638., 1670., ..., 2543., 2615., 2605.], [1802., 1827., 1907., ..., 2238., 2574., 2519.]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [1992., 2045., 2170., ..., 3259., 3767., 3800.], [2027., 2077., 2204., ..., 3008., 3374., 3356.], [2132., 2298., 2276., ..., 2572., 3123., 3148.]], [[2294., 2508., 2523., ..., 3001., 3081., 2691.], [2175., 2439., 2670., ..., 3115., 3223., 2743.], [2248., 2323., 2578., ..., 3311., 3203., 2816.], ..., [2148., 2232., 2300., ..., 3482., 3986., 3939.], [2146., 2252., 2344., ..., 3212., 3588., 3509.], [2345., 2399., 2528., ..., 2864., 3358., 3375.]], [[2310., 2502., 2571., ..., 2961., 3010., 2640.], [2280., 2497., 2731., ..., 3104., 3181., 2722.], [2281., 2397., 2626., ..., 3249., 3215., 2843.], ..., [2107., 2162., 2234., ..., 3384., 3826., 3831.], [2128., 2152., 2325., ..., 3207., 3504., 3403.], [2259., 2426., 2431., ..., 2744., 3324., 3307.]]]) - fmask(time, y, x)float64nan nan nan nan ... 1.0 1.0 1.0 1.0
- units :
- 1
- nodata :
- 0
- flags_definition :
- {'fmask': {'bits': [0, 1, 2, 3, 4, 5, 6, 7], 'values': {'0': 'nodata', '1': 'valid', '2': 'cloud', '3': 'shadow', '4': 'snow', '5': 'water'}, 'description': 'Fmask'}}
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]], [[ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]]])
- crs :
- EPSG:3577
- grid_mapping :
- spatial_ref
Using the datetime accessor
xarray allows you to easily extract additional information from the time dimension in Digital Earth Australia data. For example, we can get a list of what season each observation belongs to:
[7]:
ds.time.dt.season
[7]:
<xarray.DataArray 'season' (time: 46)>
array(['DJF', 'DJF', 'DJF', 'DJF', 'MAM', 'MAM', 'MAM', 'MAM', 'MAM',
'MAM', 'JJA', 'JJA', 'JJA', 'JJA', 'JJA', 'SON', 'SON', 'SON',
'SON', 'SON', 'SON', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF',
'MAM', 'MAM', 'MAM', 'MAM', 'MAM', 'JJA', 'JJA', 'JJA', 'JJA',
'JJA', 'JJA', 'SON', 'SON', 'SON', 'SON', 'SON', 'SON', 'DJF',
'DJF'], dtype='<U3')
Coordinates:
* time (time) datetime64[ns] 2015-01-06T00:20:26.943424 ... 2016-12...
spatial_ref int32 3577- time: 46
- 'DJF' 'DJF' 'DJF' 'DJF' 'MAM' 'MAM' ... 'SON' 'SON' 'SON' 'DJF' 'DJF'
array(['DJF', 'DJF', 'DJF', 'DJF', 'MAM', 'MAM', 'MAM', 'MAM', 'MAM', 'MAM', 'JJA', 'JJA', 'JJA', 'JJA', 'JJA', 'SON', 'SON', 'SON', 'SON', 'SON', 'SON', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'MAM', 'MAM', 'MAM', 'MAM', 'MAM', 'JJA', 'JJA', 'JJA', 'JJA', 'JJA', 'JJA', 'SON', 'SON', 'SON', 'SON', 'SON', 'SON', 'DJF', 'DJF'], dtype='<U3') - time(time)datetime64[ns]2015-01-06T00:20:26.943424 ... 2...
- units :
- seconds since 1970-01-01 00:00:00
array(['2015-01-06T00:20:26.943424000', '2015-01-22T00:20:21.686938000', '2015-02-07T00:20:18.471354000', '2015-02-23T00:20:12.433781000', '2015-03-11T00:20:02.105880000', '2015-03-27T00:19:53.920971000', '2015-04-12T00:19:46.703592000', '2015-04-28T00:19:40.864449000', '2015-05-14T00:19:24.695756000', '2015-05-30T00:19:28.951301000', '2015-06-15T00:19:41.139454000', '2015-07-01T00:19:47.228818000', '2015-07-17T00:19:57.584270000', '2015-08-02T00:20:01.251247000', '2015-08-18T00:20:08.061952000', '2015-09-03T00:20:12.861679000', '2015-09-19T00:20:21.648088000', '2015-10-05T00:20:25.198218000', '2015-10-21T00:20:27.331461000', '2015-11-06T00:20:30.948967000', '2015-11-22T00:20:32.825090000', '2015-12-08T00:20:31.424104000', '2015-12-24T00:20:32.375097000', '2016-01-09T00:20:27.829692000', '2016-01-25T00:20:28.787852000', '2016-02-10T00:20:23.428768000', '2016-02-26T00:20:17.767538000', '2016-03-13T00:20:14.830544000', '2016-03-29T00:20:05.138989000', '2016-04-14T00:19:59.638523000', '2016-04-30T00:19:59.415438000', '2016-05-16T00:19:57.493460000', '2016-06-01T00:20:03.752104000', '2016-06-17T00:20:06.101153000', '2016-07-03T00:20:15.814229000', '2016-07-19T00:20:22.384683000', '2016-08-04T00:20:25.481509000', '2016-08-20T00:20:30.301480000', '2016-09-05T00:20:36.254614000', '2016-09-21T00:20:38.199890000', '2016-10-07T00:20:41.044311000', '2016-10-23T00:20:44.869309000', '2016-11-08T00:20:43.950933000', '2016-11-24T00:20:44.489163000', '2016-12-10T00:20:41.958724000', '2016-12-26T00:20:38.145340000'], dtype='datetime64[ns]') - spatial_ref()int323577
- spatial_ref :
- PROJCS["GDA94 / Australian Albers",GEOGCS["GDA94",DATUM["Geocentric_Datum_of_Australia_1994",SPHEROID["GRS 1980",6378137,298.257222101,AUTHORITY["EPSG","7019"]],AUTHORITY["EPSG","6283"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4283"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["latitude_of_center",0],PARAMETER["longitude_of_center",132],PARAMETER["standard_parallel_1",-18],PARAMETER["standard_parallel_2",-36],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","3577"]]
- grid_mapping_name :
- albers_conical_equal_area
array(3577, dtype=int32)
Or the day of the year:
[8]:
ds.time.dt.dayofyear
[8]:
<xarray.DataArray 'dayofyear' (time: 46)>
array([ 6, 22, 38, 54, 70, 86, 102, 118, 134, 150, 166, 182, 198,
214, 230, 246, 262, 278, 294, 310, 326, 342, 358, 9, 25, 41,
57, 73, 89, 105, 121, 137, 153, 169, 185, 201, 217, 233, 249,
265, 281, 297, 313, 329, 345, 361])
Coordinates:
* time (time) datetime64[ns] 2015-01-06T00:20:26.943424 ... 2016-12...
spatial_ref int32 3577- time: 46
- 6 22 38 54 70 86 102 118 134 ... 233 249 265 281 297 313 329 345 361
array([ 6, 22, 38, 54, 70, 86, 102, 118, 134, 150, 166, 182, 198, 214, 230, 246, 262, 278, 294, 310, 326, 342, 358, 9, 25, 41, 57, 73, 89, 105, 121, 137, 153, 169, 185, 201, 217, 233, 249, 265, 281, 297, 313, 329, 345, 361]) - time(time)datetime64[ns]2015-01-06T00:20:26.943424 ... 2...
- units :
- seconds since 1970-01-01 00:00:00
array(['2015-01-06T00:20:26.943424000', '2015-01-22T00:20:21.686938000', '2015-02-07T00:20:18.471354000', '2015-02-23T00:20:12.433781000', '2015-03-11T00:20:02.105880000', '2015-03-27T00:19:53.920971000', '2015-04-12T00:19:46.703592000', '2015-04-28T00:19:40.864449000', '2015-05-14T00:19:24.695756000', '2015-05-30T00:19:28.951301000', '2015-06-15T00:19:41.139454000', '2015-07-01T00:19:47.228818000', '2015-07-17T00:19:57.584270000', '2015-08-02T00:20:01.251247000', '2015-08-18T00:20:08.061952000', '2015-09-03T00:20:12.861679000', '2015-09-19T00:20:21.648088000', '2015-10-05T00:20:25.198218000', '2015-10-21T00:20:27.331461000', '2015-11-06T00:20:30.948967000', '2015-11-22T00:20:32.825090000', '2015-12-08T00:20:31.424104000', '2015-12-24T00:20:32.375097000', '2016-01-09T00:20:27.829692000', '2016-01-25T00:20:28.787852000', '2016-02-10T00:20:23.428768000', '2016-02-26T00:20:17.767538000', '2016-03-13T00:20:14.830544000', '2016-03-29T00:20:05.138989000', '2016-04-14T00:19:59.638523000', '2016-04-30T00:19:59.415438000', '2016-05-16T00:19:57.493460000', '2016-06-01T00:20:03.752104000', '2016-06-17T00:20:06.101153000', '2016-07-03T00:20:15.814229000', '2016-07-19T00:20:22.384683000', '2016-08-04T00:20:25.481509000', '2016-08-20T00:20:30.301480000', '2016-09-05T00:20:36.254614000', '2016-09-21T00:20:38.199890000', '2016-10-07T00:20:41.044311000', '2016-10-23T00:20:44.869309000', '2016-11-08T00:20:43.950933000', '2016-11-24T00:20:44.489163000', '2016-12-10T00:20:41.958724000', '2016-12-26T00:20:38.145340000'], dtype='datetime64[ns]') - spatial_ref()int323577
- spatial_ref :
- PROJCS["GDA94 / Australian Albers",GEOGCS["GDA94",DATUM["Geocentric_Datum_of_Australia_1994",SPHEROID["GRS 1980",6378137,298.257222101,AUTHORITY["EPSG","7019"]],AUTHORITY["EPSG","6283"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4283"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["latitude_of_center",0],PARAMETER["longitude_of_center",132],PARAMETER["standard_parallel_1",-18],PARAMETER["standard_parallel_2",-36],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","3577"]]
- grid_mapping_name :
- albers_conical_equal_area
array(3577, dtype=int32)
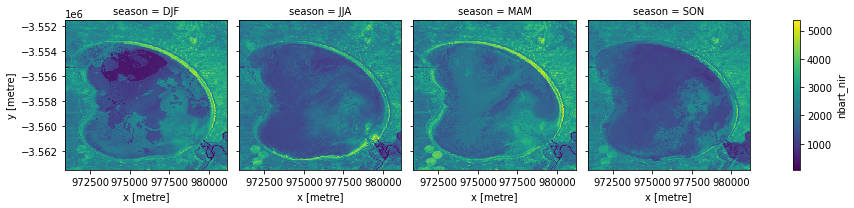
Grouping and resampling by time
xarray also provides some shortcuts for aggregating data over time. In the example below, we first group our data by season, then take the median of each group. This produces a new dataset with only four observations (one per season).
[9]:
# Group the time series into seasons, and take median of each time period
ds_seasonal = ds.groupby('time.season').median(dim='time')
# Plot the output
ds_seasonal.nbart_nir.plot(col='season', col_wrap=4)
plt.show()
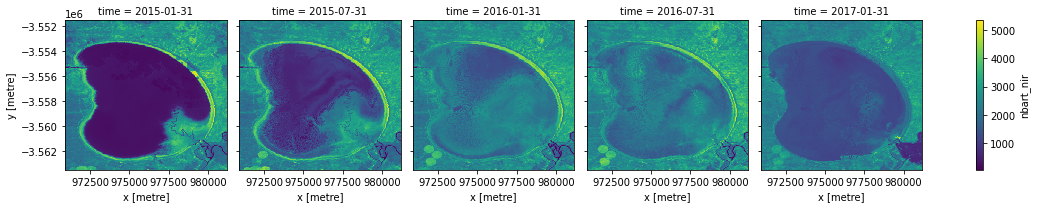
We can also use the .resample() method to summarise our dataset into larger chunks of time. In the example below, we produce a median composite for every 6 months of data in our dataset:
[10]:
# Resample to combine each 6 months of data into a median composite
ds_resampled = ds.resample(time='6m').median()
# Plot the new resampled data
ds_resampled.nbart_nir.plot(col='time')
plt.show()
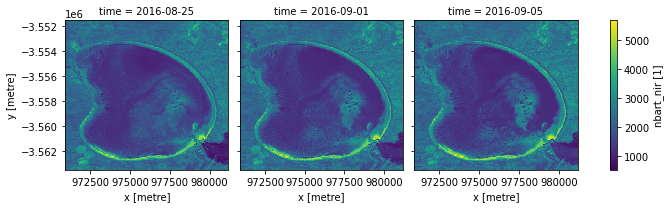
Interpolating new timesteps
Sometimes, we want to return data for specific times/dates that weren’t observed by a satellite. To estimate what the landscape appeared like on certain dates, we can use the .interp() method to interpolate between the nearest two observations.
By default, the interp() method uses linear interpolation (method='linear'). Another useful option is method='nearest', which will return the nearest satellite observation to the specified date(s).
[11]:
# New dates to interpolate data for
new_dates = ['2016-08-25', '2016-09-01', '2016-09-05']
# Interpolate Landsat values for three new dates
ds_interp = ds.interp(time=new_dates)
# Plot the new interpolated data
ds_interp.nbart_nir.plot(col='time')
plt.show()
Additional information
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Australia data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Discord chat or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
GitHub.
Last modified: December 2023
Compatible datacube version:
[12]:
print(datacube.__version__)
1.8.5
Tags
Tags: sandbox compatible, NCI compatible, dc.load, time series analysis, landsat 8, groupby, indexing, interpolating, resampling, image compositing