Determining the seasonal extent of waterbodies with Sentinel 2 
Sign up to the DEA Sandbox to run this notebook interactively from a browser
Compatibility: Notebook currently compatible with
DEA SandboxenvironmentProducts used: ga_s2am_ard_3, ga_s2bm_ard_3, ga_s2cm_ard_3
Background
The United Nations have prescribed 17 “Sustainable Development Goals” (SDGs). This notebook attempts to monitor SDG Indicator 6.6.1 - change in the extent of water-related ecosystems. Indicator 6.6.1 has 4 sub-indicators: > i. The spatial extent of water-related ecosystems > ii. The quantity of water contained within these ecosystems > iii. The quality of water within these ecosystems > iv. The health or state of these ecosystems.
This notebook primarily focuses on the first sub-indicator, spatial extents.
Description
The notebook demonstrates how to:
Load satellite data over the water body of interest
Calculate the Modified Normalized Difference Water Index (MNDWI)
Resample the time series of MNDWI to seasonal medians
Generate an animation of the water extent time series
Calculate and plot a time series of seasonal water extent (in square kilometres)
Find the minimum and maximum water extents in the time series and plot them.
Compare two nominated time periods, and plot where the waterbody extent has changed.
Getting started
To run this analysis, run all the cells in the notebook, starting with the “Load packages” cell.
Load packages
Import Python packages that are used for the analysis.
[1]:
%matplotlib inline
import datacube
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from dea_tools.bandindices import calculate_indices
from dea_tools.dask import create_local_dask_cluster
from dea_tools.datahandling import load_ard
from dea_tools.plotting import display_map, xr_animation
from IPython.display import Image
from matplotlib.colors import ListedColormap
from matplotlib.patches import Patch
Connect to the datacube
Activate the datacube database, which provides functionality for loading and displaying stored Earth observation data.
[2]:
dc = datacube.Datacube(app="Seasonal_water_extents")
Set up a Dask cluster
Dask can be used to better manage memory use and conduct the analysis in parallel. For an introduction to using Dask with Digital Earth Australia, see the Dask notebook.
Note: We recommend opening the Dask processing window to view the different computations that are being executed; to do this, see the Dask dashboard in JupyterLab section of the Dask notebook.
To activate Dask, set up the local computing cluster using the cell below.
[3]:
create_local_dask_cluster()
Client
Client-80d694c9-f4bf-11ef-82f2-ea0da03a1e87
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: /user/margaret.harrison@ga.gov.au/proxy/8787/status |
Cluster Info
LocalCluster
c7368931
| Dashboard: /user/margaret.harrison@ga.gov.au/proxy/8787/status | Workers: 1 |
| Total threads: 2 | Total memory: 12.21 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-3e15b9c0-c6ef-487e-913a-23e390f0b72c
| Comm: tcp://127.0.0.1:41673 | Workers: 1 |
| Dashboard: /user/margaret.harrison@ga.gov.au/proxy/8787/status | Total threads: 2 |
| Started: Just now | Total memory: 12.21 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:33583 | Total threads: 2 |
| Dashboard: /user/margaret.harrison@ga.gov.au/proxy/38041/status | Memory: 12.21 GiB |
| Nanny: tcp://127.0.0.1:33099 | |
| Local directory: /tmp/dask-scratch-space/worker-am5jr3f2 | |
Analysis parameters
The following cell sets the parameters, which define the area of interest and the length of time to conduct the analysis over.
The parameters are:
lat: The central latitude to analyse (e.g. -35.0958).lon: The central longitude to analyse (e.g. 149.4249).lat_buffer: The number of degrees to load around the central latitude.lon_buffer: The number of degrees to load around the central longitude.start_yearandend_year: The date range to analyse (e.g.('2017', '2020').
If running the notebook for the first time, keep the default settings below. This will demonstrate how the analysis works and provide meaningful results. The example covers Lake George near Canberra, Australia.
[4]:
# Define the area of interest
lat = -35.0958
lon = 149.4249
lat_buffer = 0.1
lon_buffer = 0.06
# Combine central lat,lon with buffer to get area of interest
lat_range = (lat - lat_buffer, lat + lat_buffer)
lon_range = (lon - lon_buffer, lon + lon_buffer)
# Define the start year and end year
start_year = "2017"
end_year = "2022"
View the area of interest on an interactive map
The next cell will display the selected area on an interactive map. The red border represents the area of interest of the study. Zoom in and out to get a better understanding of the area of interest. Clicking anywhere on the map will reveal the latitude and longitude coordinates of the clicked point.
[5]:
display_map(lon_range, lat_range)
[5]:
Load cloud-masked satellite data
The code below will create a query dictionary for our region of interest, and then load Sentinel 2 satellite data. For more information on loading data, see the Loading data notebook.
[6]:
# Create a query object
query = {
"x": lon_range,
"y": lat_range,
"resolution": (-20, 20),
"time": (start_year, end_year),
"dask_chunks": {"time": 1, "x": 2048, "y": 2048},
}
# Load Sentinel 2 data
ds = load_ard(
dc=dc,
products=["ga_s2am_ard_3", "ga_s2bm_ard_3", "ga_s2cm_ard_3"],
measurements=["green", "nbart_swir_2", "nbart_swir_3"],
cloud_mask="s2cloudless",
min_gooddata=0.9,
group_by="solar_day",
**query
)
ds
Finding datasets
ga_s2am_ard_3
ga_s2bm_ard_3
ga_s2cm_ard_3
Counting good quality pixels for each time step using s2cloudless
/env/lib/python3.10/site-packages/rasterio/warp.py:387: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
dest = _reproject(
Filtering to 133 out of 409 time steps with at least 90.0% good quality pixels
Applying s2cloudless pixel quality/cloud mask
Returning 133 time steps as a dask array
[6]:
<xarray.Dataset> Size: 1GB
Dimensions: (time: 133, y: 1177, x: 693)
Coordinates:
* time (time) datetime64[ns] 1kB 2017-01-15T00:02:42.541000 ... 20...
* y (y) float64 9kB -3.929e+06 -3.929e+06 ... -3.952e+06
* x (x) float64 6kB 1.573e+06 1.573e+06 ... 1.587e+06 1.587e+06
spatial_ref int32 4B 3577
Data variables:
green (time, y, x) float32 434MB dask.array<chunksize=(1, 1177, 693), meta=np.ndarray>
nbart_swir_2 (time, y, x) float32 434MB dask.array<chunksize=(1, 1177, 693), meta=np.ndarray>
nbart_swir_3 (time, y, x) float32 434MB dask.array<chunksize=(1, 1177, 693), meta=np.ndarray>
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refCalculate the MNDWI water index
[7]:
# Calculate MNDWI and add it to the loaded data set
ds = calculate_indices(ds=ds, index="MNDWI", collection="ga_s2_3", drop=True)
Dropping bands ['green', 'nbart_swir_2', 'nbart_swir_3']
Resample time series
Due to many factors (e.g. cloud obscuring the region, missed cloud cover in the fmask layer) the data main contain gaps or noise. Here, we will resample the data to ensure we working with a consistent time series.
To do this we resample the data to seasonal time steps using medians.
These calculations will take several minutes to complete as we will run .compute(), triggering all the tasks we scheduled above and bringing the arrays into memory.
[8]:
%%time
sample_frequency = "QS-DEC" # quarterly starting in DEC, i.e. seasonal
# Resample using medians
print("Calculating MNDWI seasonal medians...")
mndwi = ds["MNDWI"].resample(time=sample_frequency).median().compute()
# Drop any all-NA seasons
mndwi = mndwi.dropna(dim="time", how="all")
Calculating MNDWI seasonal medians...
CPU times: user 3.63 s, sys: 451 ms, total: 4.09 s
Wall time: 57.2 s
Facet plot the MNDWI time steps
[9]:
mndwi.plot.imshow(col="time", col_wrap=4, cmap="RdBu", vmax=1, vmin=-1);
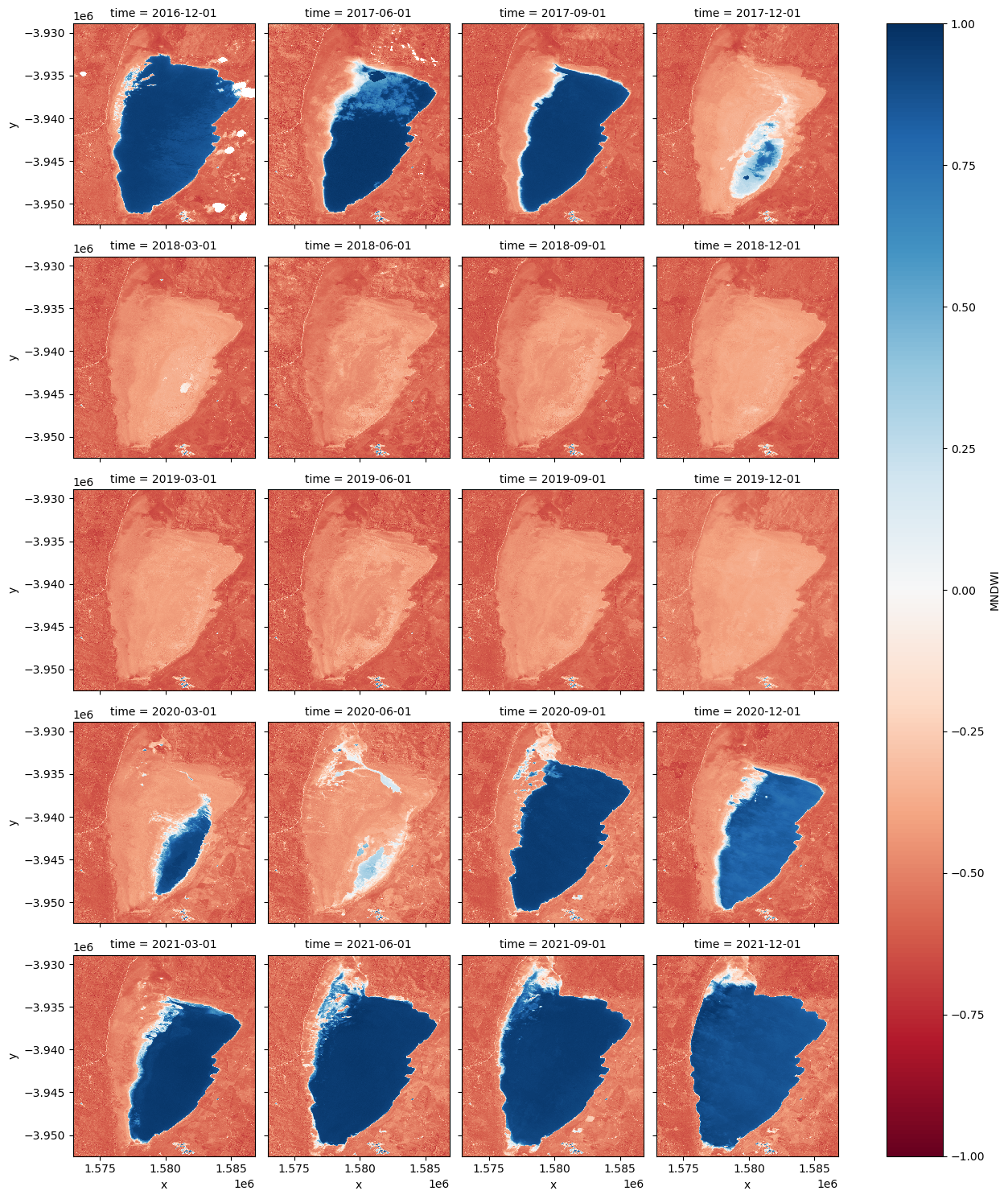
Animating time series
In the next cell, we plot the dataset we loaded above as an animation GIF, using the xr_animation function. The output_path will be saved in the directory where the script is found and you can change the names to prevent file overwrites.
[10]:
out_path = "water_extent.gif"
xr_animation(
ds=mndwi.to_dataset(name="MNDWI"),
output_path=out_path,
bands=["MNDWI"],
show_text="Seasonal MNDWI",
interval=500,
width_pixels=300,
show_colorbar=True,
imshow_kwargs={"cmap": "RdBu", "vmin": -0.5, "vmax": 0.5},
colorbar_kwargs={"colors": "black"},
)
# Plot animated gif
plt.close()
Image(filename=out_path)
Exporting animation to water_extent.gif
[10]:
<IPython.core.display.Image object>
Calculate the area per pixel
The number of pixels can be used to calculate the area of the waterbody if the pixel area is known. Run the following cell to generate the necessary constants for performing this conversion.
[11]:
pixel_length = query["resolution"][1] # in metres
m_per_km = 1000 # conversion from metres to kilometres
area_per_pixel = pixel_length**2 / m_per_km**2
Calculating the extent of water
Calculates the area of pixels classified as water (if MNDWI is > 0, then water).
[12]:
water = mndwi.where(mndwi > 0)
area_ds = water.where(np.isnan(water), 1)
ds_valid_water_area = area_ds.sum(dim=["x", "y"]) * area_per_pixel
Plot seasonal time series from the Start year to End year
[13]:
plt.figure(figsize=(18, 4))
ds_valid_water_area.plot(marker="o", color="#9467bd")
plt.title(f"Observed Seasonal Area of Water from {start_year} to {end_year}")
plt.xlabel("Dates")
plt.ylabel("Waterbody area (km$^2$)")
plt.tight_layout()
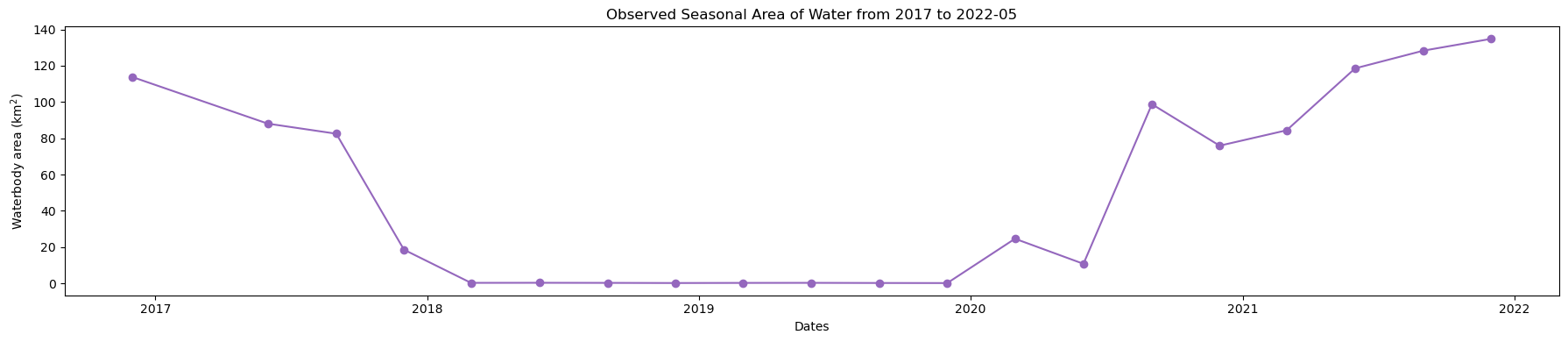
Determine minimum and maximum water extent
The next cell extracts the minimum and maximum extent of water from the dataset using the min and max functions. We then add the dates to an xarray.DataArray.
[14]:
min_water_area_date, max_water_area_date = min(ds_valid_water_area), max(
ds_valid_water_area
)
time_da = xr.DataArray(
[min_water_area_date.time.values, max_water_area_date.time.values], dims=["time"]
)
print(time_da)
<xarray.DataArray (time: 2)> Size: 16B
array(['2019-12-01T00:00:00.000000000', '2022-12-01T00:00:00.000000000'],
dtype='datetime64[ns]')
Dimensions without coordinates: time
Plot the minimum and maximum water extents
Plot the water classified pixels for the two dates where we have the minimum and maximum surface water extent.
[15]:
area_ds.sel(time=time_da).plot.imshow(col="time", col_wrap=2, figsize=(14, 6));
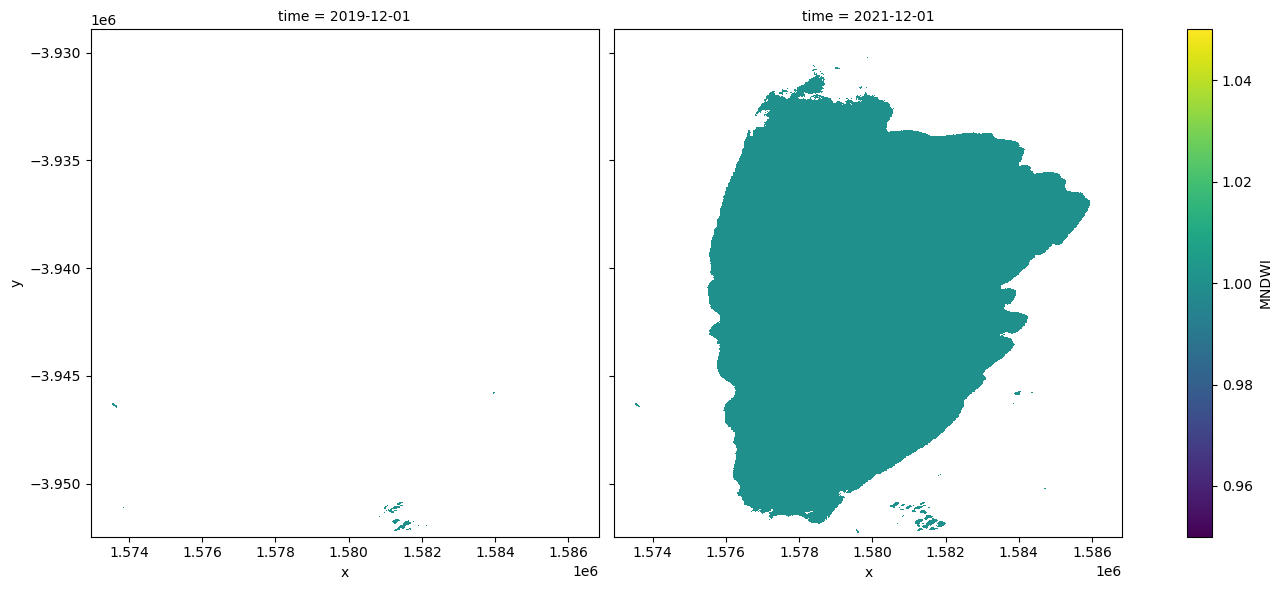
Compare two time periods
The following cells determine the maximum extent of water for two different years.
baseline_year: The baseline year for the analysisanalysis_year: The year to compare to the baseline year
[16]:
baseline_time = "2019-03-01"
analysis_time = "2020-03-01"
baseline_ds, analysis_ds = ds_valid_water_area.sel(
time=baseline_time, method="nearest"
), ds_valid_water_area.sel(time=analysis_time, method="nearest")
A new dataArray is created to store the new date from the maximum water extent for the two years
[17]:
time_da = xr.DataArray(
[baseline_ds.time.values, analysis_ds.time.values], dims=["time"]
)
Plot the water extent of the MNDWI product for the two chosen periods
[18]:
area_ds.sel(time=time_da).plot(
col="time",
col_wrap=2,
robust=True,
figsize=(10, 5),
cmap="viridis",
add_colorbar=False,
);
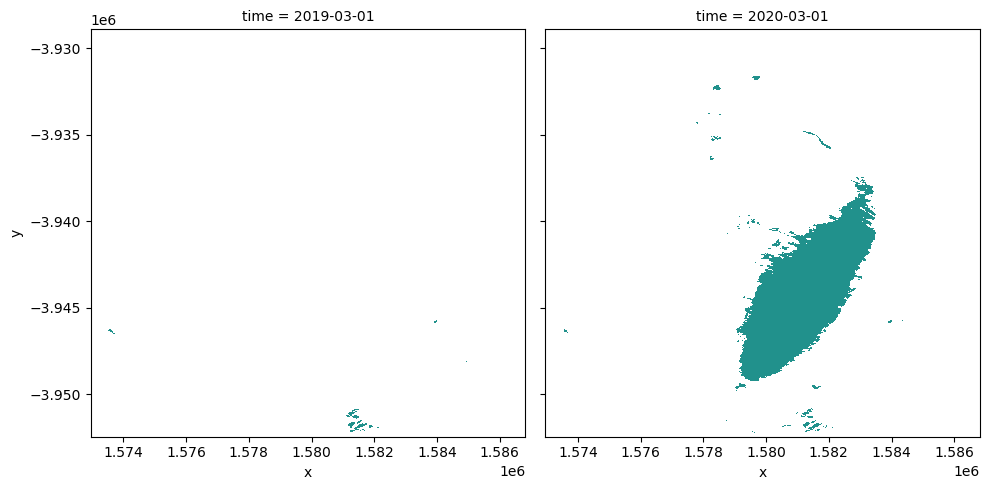
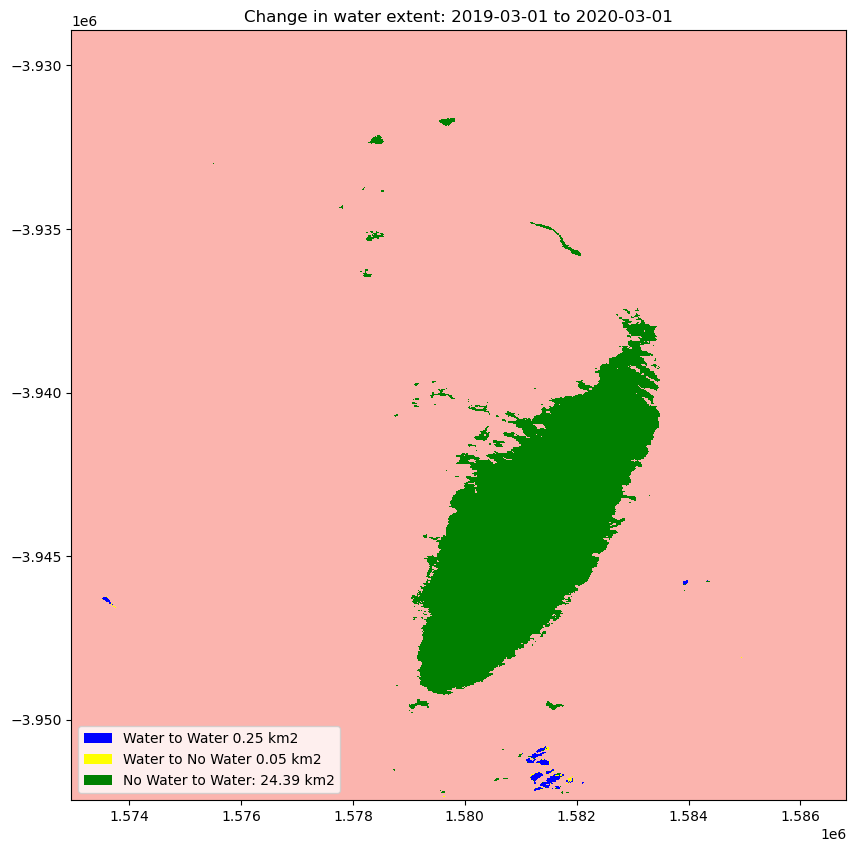
Calculating the change between two time periods
The cells below calculate the amount of water gain, loss and no change for the two time periods.
[19]:
# Extract the two time periods
ds_selected = area_ds.where(area_ds == 1, 0).sel(time=time_da)
analyse_total_value = ds_selected[1]
change = analyse_total_value - ds_selected[0]
water_appeared = change.where(change == 1)
permanent_water = change.where((change == 0) & (analyse_total_value == 1))
permanent_land = change.where((change == 0) & (analyse_total_value == 0))
water_disappeared = change.where(change == -1)
The cell below calculates the area of water extent for water_loss, water_gain, permanent water and land
[20]:
total_area = analyse_total_value.count().values * area_per_pixel
water_apperaed_area = water_appeared.count().values * area_per_pixel
permanent_water_area = permanent_water.count().values * area_per_pixel
water_disappeared_area = water_disappeared.count().values * area_per_pixel
Plot the change in water
[21]:
water_appeared_color = "Green"
water_disappeared_color = "Yellow"
stable_color = "Blue"
land_color = "Brown"
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
ds_selected[1].plot.imshow(cmap="Pastel1", add_colorbar=False, add_labels=False, ax=ax)
water_appeared.plot.imshow(
cmap=ListedColormap([water_appeared_color]),
add_colorbar=False,
add_labels=False,
ax=ax,
)
water_disappeared.plot.imshow(
cmap=ListedColormap([water_disappeared_color]),
add_colorbar=False,
add_labels=False,
ax=ax,
)
permanent_water.plot.imshow(
cmap=ListedColormap([stable_color]), add_colorbar=False, add_labels=False, ax=ax
)
plt.legend(
[
Patch(facecolor=stable_color),
Patch(facecolor=water_disappeared_color),
Patch(facecolor=water_appeared_color),
Patch(facecolor=land_color),
],
[
f"Water to Water {round(permanent_water_area, 2)} km2",
f"Water to No Water {round(water_disappeared_area, 2)} km2",
f"No Water to Water: {round(water_apperaed_area, 2)} km2",
],
loc="lower left",
)
plt.title(f"Change in water extent: {baseline_time} to {analysis_time}");
Next steps
Return to the “Analysis parameters” section, modify some values (e.g. lat, lon, start_year, end_year) and re-run the analysis. You can use the interactive map in the “View the selected location” section to find new central latitude and longitude values by panning and zooming, and then clicking on the area you wish to extract location values for. You can also use Google maps to search for a location you know, then return the latitude and longitude values by clicking the map.
Change the year also in “Compare Two Time Periods - a Baseline and an Analysis” section, (e.g. base_year, analyse_year) and re-run the analysis.
Additional information
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Australia data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Discord chat or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here).
If you would like to report an issue with this notebook, you can file one on GitHub.
Last modified: February 2025
Compatible datacube version:
[22]:
print(datacube.__version__)
1.8.19