Generating geometric median composites (geomedians) 
Sign up to the DEA Sandbox to run this notebook interactively from a browser
Compatibility: Notebook currently compatible with both the
NCIandDEA SandboxenvironmentsProducts used: ga_ls8c_ard_3
Background
Individual remote sensing images can be affected by noisy data, including clouds, cloud shadows, and haze. To produce cleaner images that can be compared more easily across time, we can create ‘summary’ images or ‘composites’ that combine multiple images into one image to reveal the median or ‘typical’ appearance of the landscape for a certain time period.
One approach is to create a geomedian. A geomedian is based on a high-dimensional statistic called the ‘geometric median’ (Small 1990), which effectively trades a temporal stack of poor quality observations for a single high-quality pixel composite with reduced spatial noise (Roberts et al. 2017). In contrast to a standard median, a geomedian
maintains the relationship between spectral bands. This allows us to conduct further analysis on the composite images just as we would on the original satellite images (e.g by allowing the calculation of common band indices like NDVI).
Description
In this notebook we will take of time series of noisy satellite images collected over a year and calculate an annual geomedian composite which is largely free of clouds and other noisy data.
Geomedian computations are expensive in terms of memory, data bandwidth, and cpu usage. The ODC has some useful functions, geomedian that allow dask to perform the computation in parallel across many threads to speed things up. In this notebook a local dask cluster is used, but the same approach should work using a larger, distributed dask cluster.
Getting started
To run this analysis, run all the cells in the notebook, starting with the “Load packages” cell.
Load packages
[1]:
%matplotlib inline
import numpy as np
import datacube
from odc.algo import to_f32, xr_geomedian, int_geomedian
import warnings
warnings.filterwarnings('ignore')
import sys
sys.path.insert(1, '../Tools/')
from dea_tools.datahandling import load_ard
from dea_tools.plotting import rgb
from dea_tools.dask import create_local_dask_cluster
Set up a dask cluster
This will help keep our memory use down and conduct the analysis in parallel. If you’d like to view the dask dashboard, click on the hyperlink that prints below the cell. You can use the dashboard to monitor the progress of calculations.
[2]:
create_local_dask_cluster()
Client
Client-b69c6283-3f21-11ef-81b2-7a43cfd2c882
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: /user/james.miller@ga.gov.au/proxy/8787/status |
Cluster Info
LocalCluster
2b1616de
| Dashboard: /user/james.miller@ga.gov.au/proxy/8787/status | Workers: 1 |
| Total threads: 2 | Total memory: 12.21 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-112398b1-0cec-41e5-a46e-a07c16ff5b07
| Comm: tcp://127.0.0.1:35075 | Workers: 1 |
| Dashboard: /user/james.miller@ga.gov.au/proxy/8787/status | Total threads: 2 |
| Started: Just now | Total memory: 12.21 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:40737 | Total threads: 2 |
| Dashboard: /user/james.miller@ga.gov.au/proxy/34885/status | Memory: 12.21 GiB |
| Nanny: tcp://127.0.0.1:35671 | |
| Local directory: /tmp/dask-scratch-space/worker-fpv7zweb | |
Connect to the datacube
[3]:
dc = datacube.Datacube(app='Generating_geomedian_composites')
Load Landsat 8 from the datacube
Here we are loading in a timeseries of cloud-masked Landsat 8 satellite images through the datacube API using the load_ard function. This will provide us with some data to work with. To limit computation and memory this example uses only three optical bands (red, green, blue).
[4]:
# Set up centre of area of interest, and area to buffer coordinates by
lat, lon = -12.25, 132.40
buffer = 0.07
# Create a reusable query
query = {
'x': (lon - buffer, lon + buffer),
'y': (lat + buffer, lat - buffer),
'time': ('2018'),
'measurements': ['nbart_green',
'nbart_red',
'nbart_blue'],
'resolution': (-30, 30),
'group_by': 'solar_day',
'output_crs': 'EPSG:3577'
}
Compared to the typical use of load_ard which by default returns data with floating point numbers containing NaN (i.e. float32), in this example we will set the dtype to 'native'. This will keep our data in its original integer data type (i.e. Int16), with nodata values marked with -999. Doing this will halve the amount of memory our data takes up, which can be extremely valuable when conducting large-scale analyses.
[5]:
# Load available data
ds = load_ard(dc=dc,
products=['ga_ls8c_ard_3'],
dask_chunks={'x': 2000, 'y': 2000},
dtype='native',
**query)
# Print output data
ds
Finding datasets
ga_ls8c_ard_3
Applying fmask pixel quality/cloud mask
Returning 23 time steps as a dask array
[5]:
<xarray.Dataset> Size: 37MB
Dimensions: (time: 23, y: 509, x: 520)
Coordinates:
* time (time) datetime64[ns] 184B 2018-01-04T01:16:45.170611 ... 20...
* y (y) float64 4kB -1.284e+06 -1.284e+06 ... -1.299e+06 -1.299e+06
* x (x) float64 4kB 3.656e+04 3.658e+04 ... 5.21e+04 5.212e+04
spatial_ref int32 4B 3577
Data variables:
nbart_green (time, y, x) int16 12MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
nbart_red (time, y, x) int16 12MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
nbart_blue (time, y, x) int16 12MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refPlot timesteps in true colour
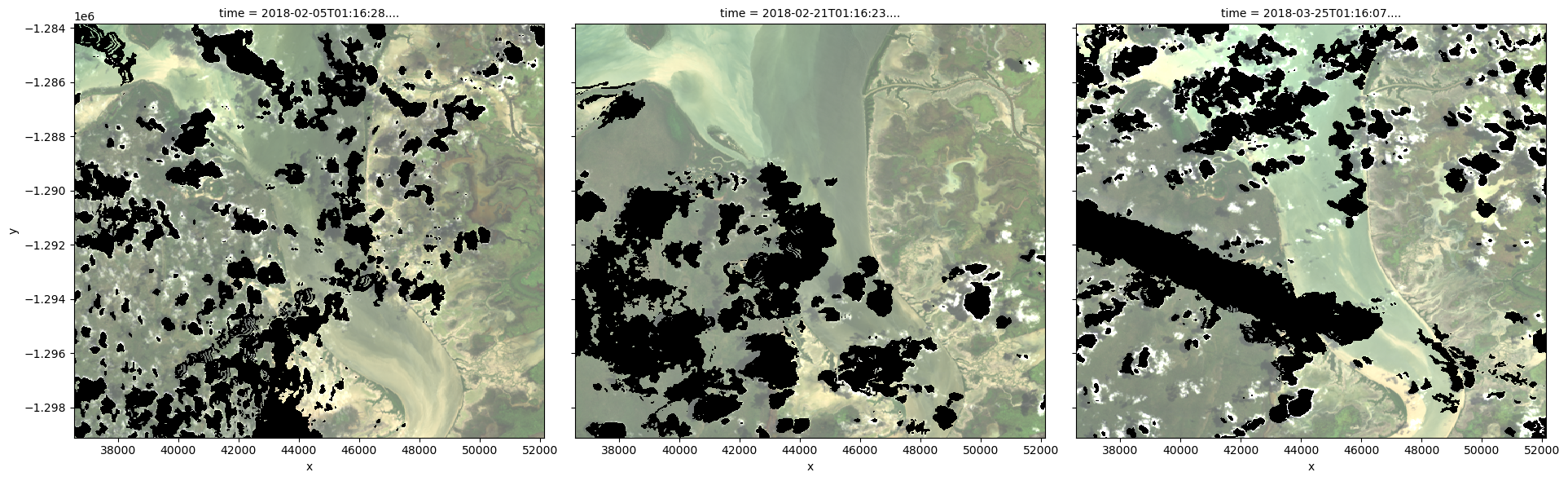
To visualise the data, use the pre-loaded rgb utility function to plot a true colour image for a series of timesteps. Black areas indicate where clouds or other invalid pixels in the image have been set to -999 to indicate no data.
The code below will plot three timesteps of the time series we just loaded.
Note: This step can be quite slow because the dask arrays being plotted must be computed first.
[6]:
# Set the timesteps to visualise
timesteps = [2, 3, 5]
# Generate RGB plots at each timestep
rgb(ds, index=timesteps)
Generate a geomedian
As you can see above, most satellite images will have at least some areas masked out due to clouds or other interference between the satellite and the ground. Running the int_geomedian function will generate a geomedian composite by combining all the observations in our xarray.Dataset into a single, complete (or near complete) image representing the geometric median of the time period.
Note: Because our data was lazily loaded with
dask, the geomedian algorithm itself will not be triggered until we call the.compute()method in the next step.
[7]:
geomedian = int_geomedian(ds)
Run the computation
The .compute() method will trigger the computation of the geomedian algorithm above. This will take about a few minutes to run; view the dask dashboard to check the progress.
[8]:
%%time
geomedian = geomedian.compute()
/env/lib/python3.10/site-packages/rasterio/warp.py:344: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
_reproject(
CPU times: user 1.45 s, sys: 291 ms, total: 1.74 s
Wall time: 49.6 s
If we print our result, you will see that the time dimension has now been removed and we are left with a single image that represents the geometric median of all the satellite images in our initial time series:
[9]:
geomedian
[9]:
<xarray.Dataset> Size: 2MB
Dimensions: (y: 509, x: 520)
Coordinates:
* y (y) float64 4kB -1.284e+06 -1.284e+06 ... -1.299e+06 -1.299e+06
* x (x) float64 4kB 3.656e+04 3.658e+04 ... 5.21e+04 5.212e+04
Data variables:
nbart_green (y, x) int16 529kB 1353 1377 1372 1353 ... 1059 1064 1030 1026
nbart_red (y, x) int16 529kB 930 961 951 919 915 ... 1365 1381 1338 1320
nbart_blue (y, x) int16 529kB 1065 1091 1089 1072 1072 ... 756 758 737 734
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refPlot the geomedian composite
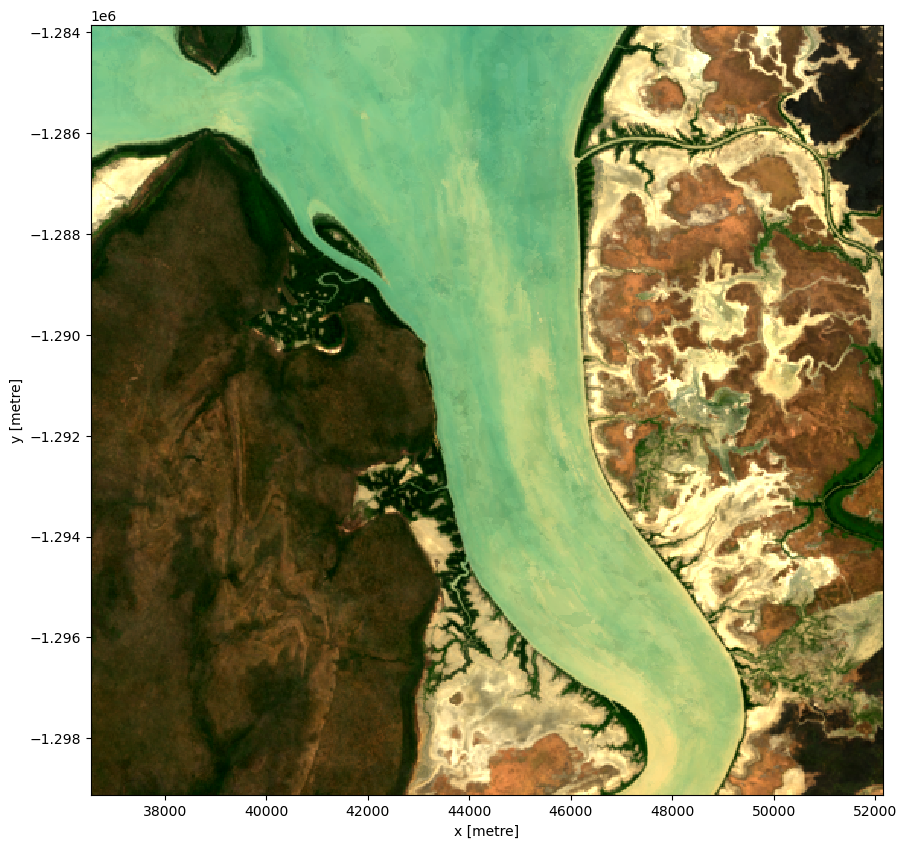
Plotting the result, we can see that the geomedian image is much more complete than any of the individual images. We can also use this data in downstream analysis as the relationships between the spectral bands are maintained by the geometric median statistic.
[10]:
# Plot the result
rgb(geomedian, size=10)
Running geomedian on grouped or resampled timeseries
mean and median are run on timeseries data that has been resampled or grouped.First we will split the timeseries into the desired groups. Resampling can be used to create a new set of times at regular intervals:
grouped = da_scaled.resample(time=1M)'nD'- number of days (e.g.'7D'for seven days)'nM'- number of months (e.g.'6M'for six months)'nY'- number of years (e.g.'2Y'for two years)
Grouping works by looking at part of the date, but ignoring other parts. For instance, 'time.month' would group together all January data together, no matter what year it is from.
grouped = da_scaled.groupby('time.month')'time.day'- groups by the day of the month (1-31)'time.dayofyear'- groups by the day of the year (1-365)'time.week'- groups by week (1-52)'time.month'- groups by the month (1-12)'time.season'- groups into 3-month seasons:'DJF'December, January, February'MAM'March, April, May'JJA'June, July, August'SON'September, October, November
'time.year'- groups by the year
Here we will resample into three four-monthly groups ('4M'), with the group starting at the start of the month (represented by the 'S' at the end).
[11]:
grouped = ds.resample(time='4MS')
grouped
[11]:
DatasetResample, grouped over '__resample_dim__'
3 groups with labels 2018-01-01, ..., 2018-09-01.
Now we will apply the int_geomedian function to each resampled group using the map method.
Instead of calling int_geomedian(ds) on the entire array, we pass the int_geomedian function to map to apply it separately to each resampled group.
[12]:
geomedian_grouped = grouped.map(int_geomedian)
We can now trigger the computation, and watch progress using the dask dashboard.
[13]:
%%time
geomedian_grouped = geomedian_grouped.compute()
CPU times: user 1.78 s, sys: 322 ms, total: 2.11 s
Wall time: 1min 1s
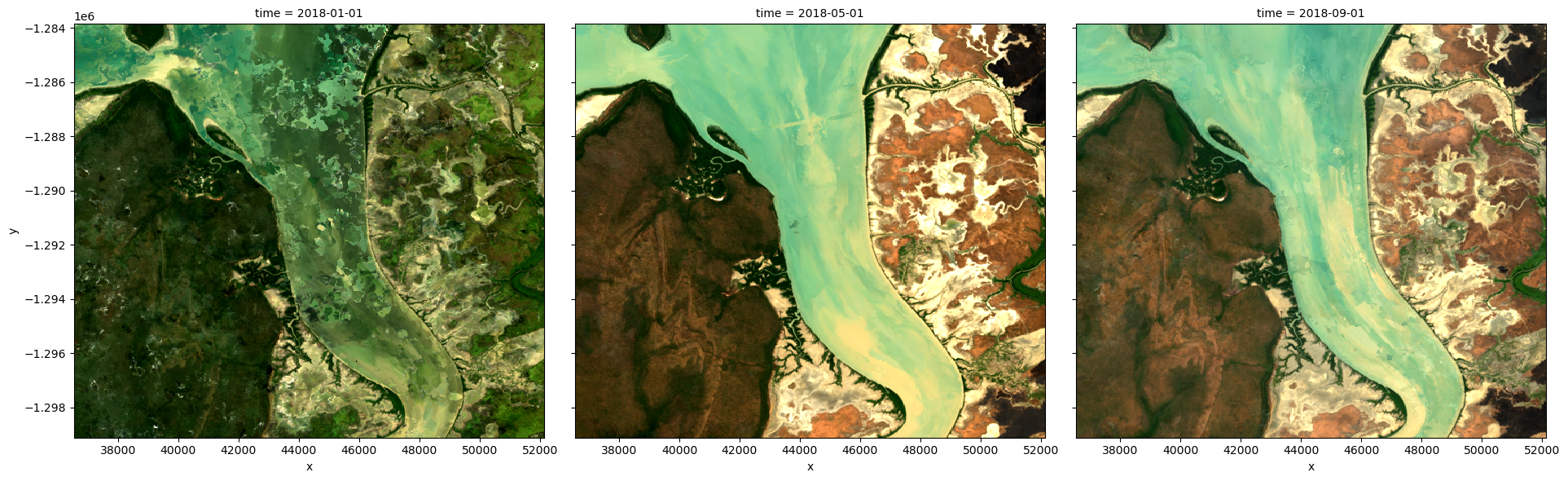
We can plot the output geomedians, and see the change in the landscape over the year:
[14]:
rgb(geomedian_grouped, col='time', col_wrap=4)
Advanced: Geomedian on float arrays
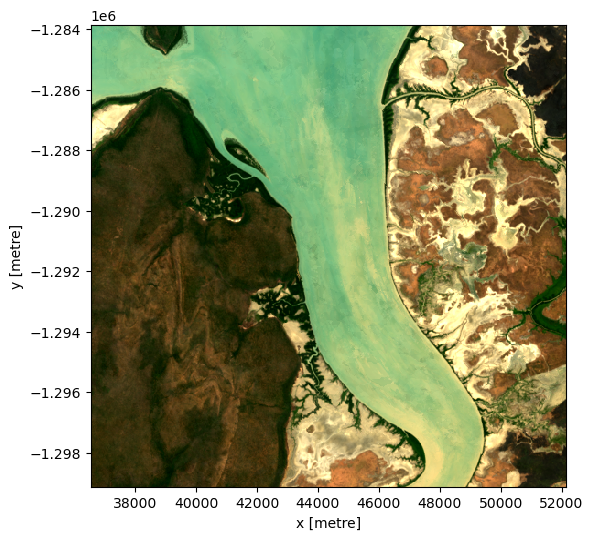
The ODC has a useful function, geomedian that allows for calcuating geomedians on a xarray.Dataset (as well as xr.DataArrays and numpy arrays) with a float datatype.
To demonstrate this we will reload our dataset using load_ard, but this time not set dtype='native'. This will return our dataset in dtype Float32.
[15]:
# Load available data
ds = load_ard(dc=dc,
products=['ga_ls8c_ard_3'],
dask_chunks={'x': 2000, 'y': 2000},
**query)
# Print output data
ds
Finding datasets
ga_ls8c_ard_3
Applying fmask pixel quality/cloud mask
Returning 23 time steps as a dask array
[15]:
<xarray.Dataset> Size: 73MB
Dimensions: (time: 23, y: 509, x: 520)
Coordinates:
* time (time) datetime64[ns] 184B 2018-01-04T01:16:45.170611 ... 20...
* y (y) float64 4kB -1.284e+06 -1.284e+06 ... -1.299e+06 -1.299e+06
* x (x) float64 4kB 3.656e+04 3.658e+04 ... 5.21e+04 5.212e+04
spatial_ref int32 4B 3577
Data variables:
nbart_green (time, y, x) float32 24MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
nbart_red (time, y, x) float32 24MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
nbart_blue (time, y, x) float32 24MB dask.array<chunksize=(1, 509, 520), meta=np.ndarray>
Attributes:
crs: EPSG:3577
grid_mapping: spatial_refNote: xr_geomedian has several parameters we can set that will control its functionality:
- Setting
num_thread=1will disable the internal threading and instead allow parallelisation withdask.Theepsparameter controls the number of iterations to conduct; a good default is1e-7. For numerical stability, it can also be advisable to scale surface reflectance float values in the dataset to 0-1 (instead of 0-10,000 as is the default for LS8). We can do this by using the helper functions
to_f32. We do this in the code cell below before we compute the geomedian. Note, this is not an essential step.
[16]:
# Scale the values using the f_32 util function
sr_max_value = 10000 # maximum SR value in the loaded product
scale, offset = (1 / sr_max_value, 0) # differs per product, aim for 0-1 values in float32
ds_scaled = to_f32(ds,
scale=scale,
offset=offset)
[17]:
geomedian = xr_geomedian(ds_scaled,
num_threads=1,
eps=1e-7,
).compute()
[18]:
rgb(geomedian)
Additional information
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Australia data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Discord chat or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
GitHub.
Last modified: July 2024
Compatible datacube version:
[19]:
print(datacube.__version__)
1.8.18
Tags
Tags: NCI compatible, sandbox compatible, landsat 8, load_ard, rgb, geomedian, image compositing, int_geomedian, xr_geomedian